# R v4.5.2
library(assertthat) # CRAN v0.2.1
library(cli) # CRAN v3.6.5
library(ggtext) # CRAN v0.1.2
library(taxizedb) # CRAN v0.3.2
library(tidyverse) # CRAN v2.0.0
library(words) # CRAN v1.0.1
library(here) # CRAN v1.0.2That’s a question Franz Anthony posted last year on Bluesky. It was late at night in my timezone and I was sleepily scrolling, I think, but this did not stop me from bolting awake, grabbing my laptop, and trying to solve it right then. It is after all:
- a biology-related question
- that can be answered with relatively simple coding, resulting in
- some very useless trivia1.
Which is, as anyone who knows me enough can tell you, also known as “extremely my niche”.
At the time, it took me a few tries to get it right, and my code was messy and not fit to show (remember, “late at night”?2), but I managed to find the answer(s).
Flash-forward to earlier this month, and this other Bluesky post, which made me remember that, and then made me think that maybe I should try to clean it and write it up properly? This could even make a blog post? So here we are.

Step 1: We need names
There are a few databases of species names out there, depending on what you need, but the Catalogue of Life (https://www.catalogueoflife.org/) is meant to be the most comprehensive. We could manually download the database from their website and work from that, but why would we when there are a bunch of R packages specifically written to do the same thing and make it painless? So we’re going to use taxizedb to do that.
Unless you already have a version of the database created by taxizedb on your computer, you first need to create one:
db_download_col() # by default: does nothing if DB already downloadedOn my laptop it takes a couple minutes to download, unzip and build the database3. When it’s done, we can have a look:
CoL <- src_col() |> tbl("taxa")
glimpse(CoL)Rows: ??
Columns: 22
Database: sqlite 3.51.1 [C:\Users\maxim\AppData\Local\cache\R\taxizedb\col.sqlite]
$ taxonID <chr> "9HCZJ", "4RMCK", "6Y3HY", "6X2FN", "6VG44", …
$ parentNameUsageID <chr> "7NZCN", "77B5", NA, "63R7G", NA, "9CL82", NA…
$ acceptedNameUsageID <chr> NA, NA, "38LCS", NA, "4H6R9", NA, "BQGDS", "4…
$ originalNameUsageID <chr> "ttMKNNs9U8868GpWbNZ-v2", NA, NA, NA, NA, NA,…
$ scientificNameID <chr> "---6DZbhwZdd3GzHPGmEg2", "---7IxQ-e98g90AiUv…
$ datasetID <int> 1130, 1141, 1106, 1029, 2304, 2304, 2299, 114…
$ taxonomicStatus <chr> "accepted", "accepted", "synonym", "accepted"…
$ taxonRank <chr> "species", "species", "species", "species", "…
$ scientificName <chr> "Chondrula tchetchenica Steklov, 1962", "Rauv…
$ scientificNameAuthorship <chr> "Steklov, 1962", "A. S. Rao", "Michelin, 1862…
$ notho <int> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ genericName <chr> "Chondrula", "Rauvolfia", "Savignya", "Pyrnus…
$ infragenericEpithet <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ specificEpithet <chr> "tchetchenica", "leptophylla", "frappieri", "…
$ infraspecificEpithet <chr> NA, NA, NA, NA, NA, NA, NA, "digitotuberosum"…
$ cultivarEpithet <int> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ nameAccordingTo <int> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ namePublishedIn <chr> NA, "Rao, A. S. (1956). In: Ann. Missouri Bot…
$ nomenclaturalCode <chr> "ICZN", "ICN", "ICZN", "ICZN", "ICN", "ICN", …
$ nomenclaturalStatus <chr> "nomen validum", NA, NA, NA, "nomen illegitim…
$ taxonRemarks <chr> NA, NA, NA, NA, NA, "S. Africa", NA, NA, NA, …
$ references <chr> "https://www.molluscabase.org/aphia.php?p=tax…The question was “What’s the highest score we could get in Scrabble if we play a taxonomically valid genus/species name?” So I’m going to restrict our search to taxa that are currently considered accepted, and search both genus names alone, and full species names. As I just wrote, taxizedb creates a persistent database rather than classical in-memory data tables. Here this changes almost nothing to the normal dplyr select-and-filter pipeline, we just need to add a collect() step at the end to pull the result of the query back in-memory:
Extract list of accepted genus names
genus_names <- CoL |>
filter(
!is.na(genericName) &
taxonomicStatus == "accepted"
) |>
select(genericName) |>
distinct() |>
collect() |>
pull(genericName)Extract list of accepted species names
species_names <- CoL |>
filter(
!is.na(genericName) &
!is.na(specificEpithet) &
taxonRank == "species" &
taxonomicStatus == "accepted"
) |>
mutate(specificName = paste(genericName, specificEpithet, sep = " ")) |>
select(specificName) |>
distinct() |>
collect() |>
pull(specificName)The R function distinct() in the code blocks above is meant to filter out duplicates. For genus names, obviously we’re going to have genera with multiple species in the database, but should we expect duplicates in the species names table too? Yes, because different parts of the Tree of Life are governed by different taxonomic codes4, and for instance there’s no rule against using a name for plants that’s already used for animals5.
length(genus_names)[1] 204147length(species_names)[1] 2057698So this gives us: 204147 genus names and 2057698 species names to check. Whew!
Step 2: We need to be able to score these names
To do that, we first need a list of the point values of each letter, and a list of how many tiles there are for each letter in a Scrabble set. Because yes, I am going to do this properly. I’m not going to just count the point values of all the names and declare the biggest scorer the winner6, I am going to make sure the winning names can fit on a 15 by 15 Scrabble board, and that there are enough tiles to play them. The letter distributions are available on Wikipedia, let’s just do English for this one.
Given a vector of names, what we need to do is:
- for each name, count how many of each letter it has
- check how many tiles it uses (with and without using blank tiles to make the space between genus and species)
- combine these to the Scrabble letter distribution
- score the name unless there’s not enough letters
- select the best-scoring name among the names that fit on the board.
I’ve encoded all these steps into the function below (which can be used for any vector of alphabetical strings, not just the taxonomic names produced here):
Scrabble scoring function
scrabble_score <- function(strings,
return_all_scores = TRUE # if FALSE, returns only top scorers
) {
if (!require(cli)) {
stop("package cli is needed and not installed")
}
if (!require(dplyr)) {
stop("tidyverse package dplyr is needed and not installed")
}
if (!require(purrr)) {
stop("tidyverse package purrr is needed and not installed")
}
if (!require(stringr)) {
stop("tidyverse package stringr is needed and not installed")
}
if (!require(tibble)) {
stop("tidyverse package tibble is needed and not installed")
}
if (!require(tidyr)) {
stop("tidyverse package tidyr is needed and not installed")
}
if (!require(assertthat)) {
stop("package assertthat is needed and not installed")
}
assert_that(is.vector(strings))
assert_that(is.character(strings))
assert_that(is.logical(return_all_scores))
# EN letters distributions sourced from wikipedia page on 2026-01-10
tiles <- tribble(
~character, ~value, ~frequency,
"a", 1, 9,
"b", 3, 2,
"c", 3, 2,
"d", 2, 4,
"e", 1, 12,
"f", 4, 2,
"g", 2, 3,
"h", 4, 2,
"i", 1, 9,
"j", 8, 1,
"k", 5, 1,
"l", 1, 4,
"m", 3, 2,
"n", 1, 6,
"o", 1, 8,
"p", 3, 2,
"q", 10, 1,
"r", 1, 6,
"s", 1, 4,
"t", 1, 6,
"u", 1, 4,
"v", 4, 2,
"w", 4, 2,
"x", 8, 1,
"y", 4, 2,
"z", 10, 1,
" ", 0, 2
)
# score each name:
# put names to all lowercase
# count how many of each letter in name
# join with chosen tile distribution, sum
# names for which there are not enough tiles score NA
scores <- tibble(string = str_to_lower(unique(strings))) |>
mutate(
length = str_length(string),
length_nospace = str_length(str_remove_all(string, " ")),
tiles = list(tiles)
) |>
mutate(score = map2(
.x = tiles,
.y = string,
.f = function(.x, .y) {
.x |>
mutate(string = .y) |>
mutate(n = str_count(string, character)) |>
mutate(
effective_n = case_when(
n > frequency ~ NA_integer_,
# inject NA in the calculation if a string has more of a letter than there are tiles
TRUE ~ n
)
) |>
summarize(score = sum(effective_n * value))
},
.progress = list(
format = "scoring name {cli::pb_current}/{cli::pb_total} {cli::pb_bar} | ETA: {cli::pb_eta}"
)
)) |>
unnest(score) |>
select(string, length, length_nospace, score)
# find the best scorers
topscore_classic <- scores |>
filter(length <= 15) |>
filter(score == max(score, na.rm = TRUE))
topscore_squish_spaces <- scores |>
filter(length_nospace <= 15) |>
filter(score == max(score, na.rm = TRUE))
# return best scorers
if (return_all_scores == TRUE) {
to_return <- list(
topscore_classic = topscore_classic,
topscore_squish_spaces = topscore_squish_spaces,
tiles = tiles,
scores = scores
)
} else {
to_return <- list(
topscore_classic = topscore_classic,
topscore_squish_spaces = topscore_squish_spaces,
tiles = tiles
)
}
return(to_return)
}What is the highest scoring genus then7?
genus_scores <- scrabble_score(genus_names)genus_scores$topscore_classic# A tibble: 1 × 4
string length length_nospace score
<chr> <int> <int> <dbl>
1 xochiquetzallia 15 15 45The genus name that would get you the highest score (not counting bonuses) out of an English-language Scrabble game is Xochiquetzallia. It’s a recently described (2020) plant genus, that groups a bunch of species previously classified into 2 other genera. They are rare and/or poorly recorded flowering plants, only found in one region of south Mexico, and with less than 100 observations in GBIF all species combined. To cite the authors:
This genus is named in honor of the goddess of Aztec flowers, in Nahuatl “Xōchiquetzalli” (beautiful flower) “xṓchitl” (flower), “quétzalli” (beautiful).

And what are the highest scoring species?
There’s an extra wrinkle here: do you insist on using a blank tile to mark the space between genus and species name, or are you OK with squishing them together on the board? The function lets you check both:
species_scores <- scrabble_score(species_names)species_scores$topscore_classic# A tibble: 1 × 4
string length length_nospace score
<chr> <int> <int> <dbl>
1 ixchela viquezi 15 14 47Because we’re having a theme going apparently, the highest scoring species if we’re insisting on having a blank tile between genus and species8 also has a genus name based on a Mesoamerican deity: Ixchela viquezi. It’s a spider in the family Pholcidae, the same family as the common cellar spiders; there’s again not that many records of the genus out there (though more than Xochiquetzallia), and I haven’t managed to find reliably identified and openly accessible photos of live spiders of that species, only others in the genus:

species_scores$topscore_squish_spaces# A tibble: 1 × 4
string length length_nospace score
<chr> <int> <int> <dbl>
1 anthrax jazykovi 16 15 51If you’re OK with removing the space between genus and species to get one more tile9, then I guess the top species then is Anthraxjazykovi… I mean Anthrax jazykovi.
Completely unrelated to the anthrax you may be thinking about, it’s actually a genus of bee flies, which for once in this post is actually quite common and widespread. This specific species though is even harder to track down online, with no records in GBIF to date, and only a few checklists and a short description in the Biodiversity Heritage Library.

What about the other names?
I (my laptop) just spent a few hours scoring all these names; it would kinda be a waste not to have a look, after all.
First, most genus names are playable (= not too long + enough tiles)
N_playable_genus <- genus_scores$scores |>
filter(!is.na(score) & length <= 15) |>
count() |>
pull(n)
N_playable_genus[1] 189778(that’s about 93% of the names).
But that’s not the case at all for species names, with the vast majority of names being unplayable, to the point that there’s not that many more playable species names than there are genus names
N_playable_species <- species_scores$scores |>
filter(!is.na(score) & length <= 15) |>
count() |>
pull(n)
N_playable_species[1] 208009(that’s only about 10.1% of the names10).
We can do a couple plots to look at the names that are valid. Let’s start by preparing a few things:
Getting and scoring Scrabble-allowed English words
data("words")
words_scores <- scrabble_score(words$word)Preparing results for plots
summary_words <- words_scores$scores |>
mutate(nletters = length) |>
group_by(nletters) |>
summarize(
mean_score_words = mean(score, na.rm = TRUE),
median_score_words = median(score, na.rm = TRUE)
)
## note to self: it has NAs because you can use the blank tiles to play words with more letters than in the bag in normal play
genus_outliers <- genus_scores$scores |>
mutate(nletters = length) |>
filter(nletters <= 15 & !is.na(score)) |>
group_by(nletters) |>
filter(nletters %in% c(2, 4, 8, 10, 15) & score == max(score)) |>
mutate(string = paste0("_", str_to_sentence(string), "_"))
genus_scores_summarized <- genus_scores$scores |>
mutate(nletters = length) |>
filter(nletters <= 15 & !is.na(score)) |>
group_by(nletters) |>
mutate(relative_rank = rank(score) / length(score)) |>
group_by(nletters, relative_rank, score) |>
count()
species_outliers <- species_scores$scores |>
mutate(nletters = length_nospace) |>
filter(nletters <= 14 & !is.na(score)) |>
group_by(nletters) |>
filter(nletters %in% c(4, 6, 9, 10, 12, 14) & score == max(score)) |>
mutate(string = paste0("_", str_to_sentence(string), "_"))
species_scores_summarized <- species_scores$scores |>
mutate(nletters = length_nospace) |>
filter(nletters <= 14 & !is.na(score)) |>
group_by(nletters) |>
mutate(relative_rank = rank(score) / length(score)) |>
group_by(nletters, relative_rank, score) |>
count()A custom plot function
make_summary_plot <- function(
scores_summarized,
words_summarized,
outliers,
breaks_bubble_legend
) {
ggplot(scores_summarized) +
geom_path(
data = words_summarized,
aes(nletters, median_score_words),
linewidth = 1
) +
geom_richtext(
data = outliers,
aes(nletters, score, label = string),
hjust = "right", vjust = "bottom",
fill = "cornsilk", label.color = "orange"
) +
geom_point(data = outliers, aes(nletters, score), col = "orange", size = 2.5) +
geom_point(
aes(
nletters,
group = nletters,
score,
fill = relative_rank,
size = n
),
pch = 21
) +
geom_segment(x = 13.5, xend = 13.5, y = 2, yend = 20.5) +
geom_richtext(
x = 13.5, y = 3, size = 2.5,
label = "Score of the median<br>Scrabble-allowed English<br>word of the same length",
hjust = 0.5,
fill = "grey95"
) +
labs(
size = "Frequency:",
fill = "For a given length:"
) +
scale_fill_distiller(
breaks = c(0, 0.5, 1),
labels = c("worst score", "median score", "best score"),
type = "div", palette = "PuOr",
limits = c(0, 1)
) +
scale_y_continuous("Score") +
scale_x_continuous("Number of letters", breaks = 1:15) +
scale_size(
breaks = breaks_bubble_legend,
limits = c(min(breaks_bubble_legend), max(breaks_bubble_legend))
) +
coord_cartesian(xlim = c(1, 15), ylim = c(1, 50)) +
theme_bw() +
theme(
panel.grid.major.x = element_blank(),
panel.grid.minor.x = element_blank(),
legend.title = element_markdown(size = 10),
legend.text = element_text(size = 9)
)
}And now let’s look:
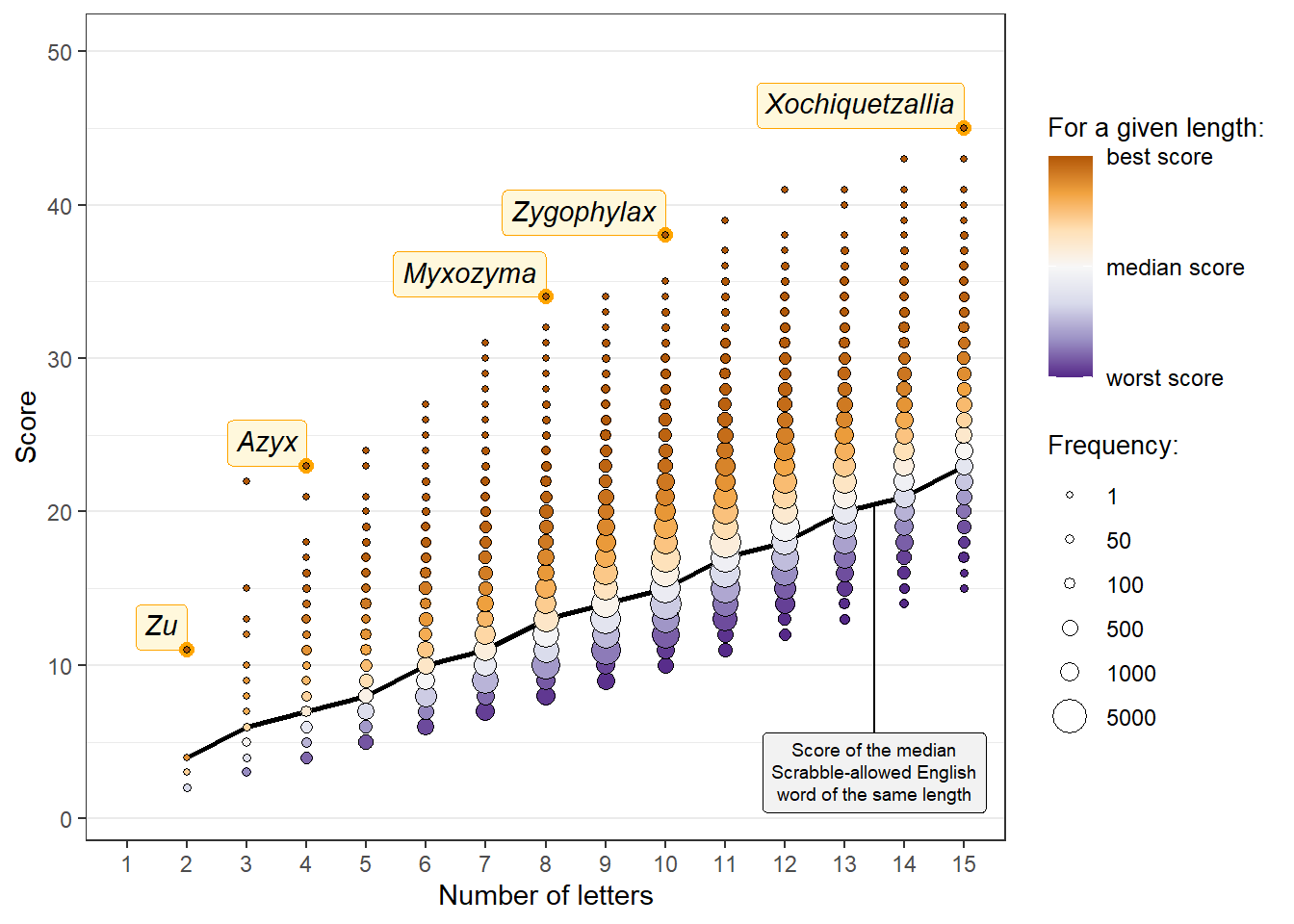
make_summary_plot(
scores_summarized = genus_scores_summarized,
words_summarized = summary_words,
outliers = genus_outliers,
breaks_bubble_legend = c(1, 50, 100, 500, 1000, 5000)
)
make_summary_plot(
scores_summarized = species_scores_summarized,
words_summarized = summary_words,
outliers = species_outliers,
breaks_bubble_legend = c(1, 500, 1000, 5000, 10000, 15000)
)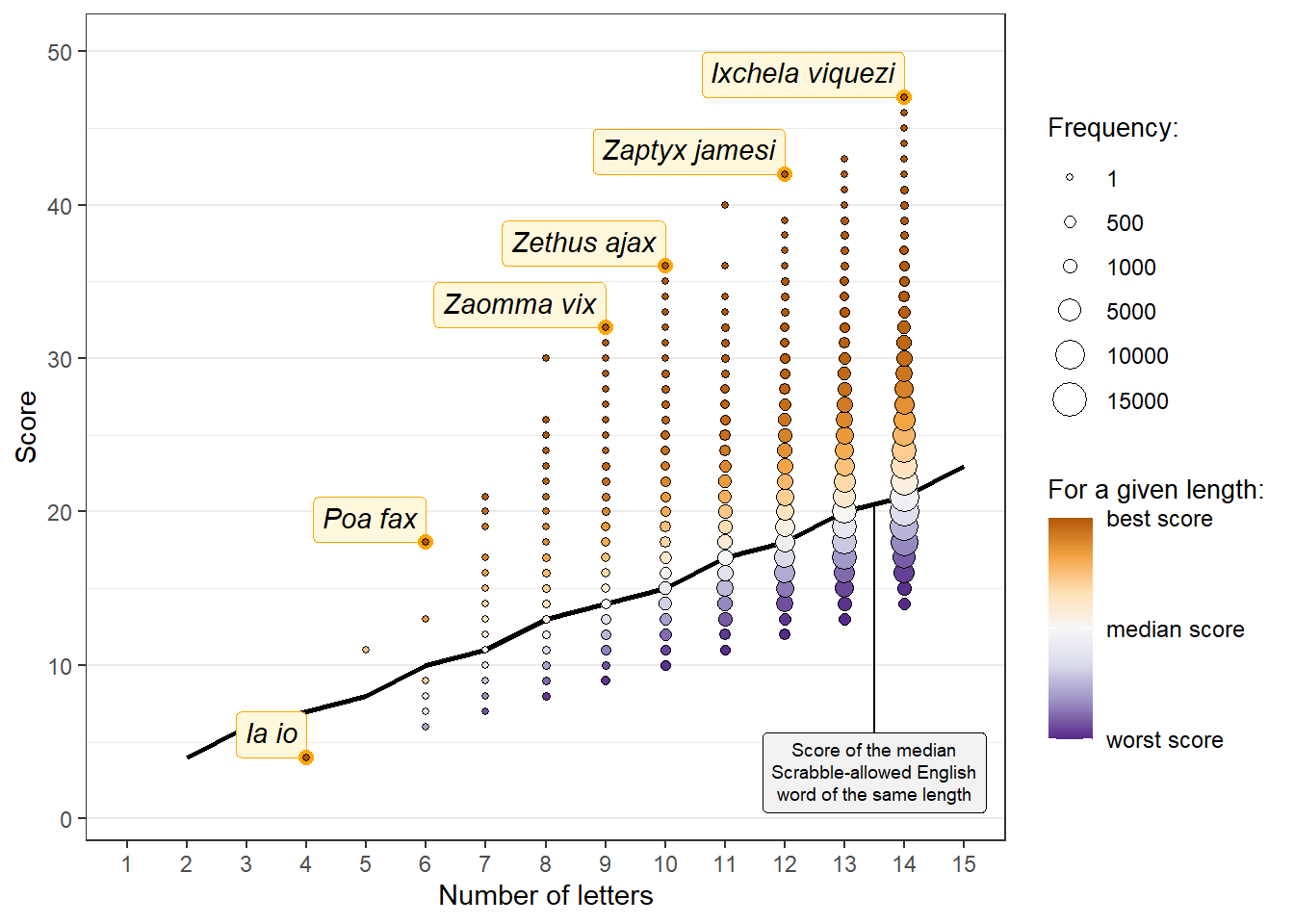
So:
For any given number of letters, the median genus or species names score about the same as the median Scrabble-allowed English word11
Among genera, a few top-scoring names for their length (without ex-aequos) include Zu (a genus of ribbonfishes), Azyx (moths), Myxozyma (fungi) or Zygophylax (cnidarians)
For species, we have Ia io (bats), and between it and Ixchela viquezi, a bunch of names I’m pretty sure I’ve seen in Star Wars, like Poa fax (a grass), or Zaomma vix and Zethus ajax (two wasps)12.
So what was the point?
Absolutely none whatsoever, I thought this was clear at the beginning?
I did learn a little about a rare genus of pretty Mexican flowers along the way, though.
Footnotes
And by “useless”, I do mean “useless”. This is not even the kind of biology trivia you can use to wow/gross out people at parties, for instance.↩︎
Please don’t remember I literally just wrote “relatively simple coding”↩︎
It takes about 2Gb on disk. If needed, it can be easily deleted directly from R; see https://docs.ropensci.org/taxizedb/reference/tdb_cache.html↩︎
https://en.wikipedia.org/wiki/Nomenclature_codes#Codification_of_scientific_names↩︎
I learned while writing this that these are called hemihomonyms https://en.wikipedia.org/wiki/Homonym_(biology)#Hemihomonyms↩︎
https://en.wikipedia.org/wiki/Myxococcus_llanfairpwllgwyngyllgogerychwyrndrobwllllantysiliogogogochensis↩︎
Running this takes a while; a few ms per name on my laptop, but there are after all many many names to check each time. It can probably be made faster, but I didn’t really try (I gave more thought to making sure it had a meaningful progress bar, tbh).↩︎
The correct way↩︎
I will judge you though↩︎
As I’ve made clear in the last two footnotes, obviously I’m assuming the scenario where you use a blank tile for the space. For the record, the other scenario adds about 130000 species, but that still leaves the vast majority of species names unplayable↩︎
Based on the
wordsR package https://cran.r-project.org/web/packages/words/index.html↩︎Zaptyx jamesi (a snail) feels more like Zaphod Beeblebrox, somehow↩︎
Reuse
Citation
@online{dahirel2026,
author = {Dahirel, Maxime},
title = {What’s the Highest Score We Could Get in {Scrabble} If We
Play a Taxonomically Valid Genus/Species Name?},
date = {2026-01-26},
url = {https://mdahirel.github.io/posts/2026-01-26-taxonomic-scrabble/},
langid = {en}
}